



A.I. Technology is Changing Rapidly!! (Molecular Genetics and Sexed Semen)
David Thorbahn, Select Sires, Inc., explains where the practise of artificial insemination is heading in the future. His essay, taken from the proceedings 45th Florida Dairy Production Conference, was published by the University of Florida, IFAS Extension.
New Tools Coming In Bovine Genetic Development
The United States has been a leader in the implementation and development of new genetic tools to advance dairy and beef populations. Today, more dairy genetics are exported to countries from the U.S. than from any other major dairy producing country in the world. I would like to give you a brief history of the advances in genetic improvement and introduce to you the new technology in molecular genetics. Then, I would like to explain some of the work currently underway and close by providing you insight into what tools you can expect to improve the health and production of your herd.
Genetic improvement started in the mid-1800s when European breed societies were formed to track parentage and record characteristics in various dairy and beef breeds. The U.S. followed suit a few years later. In addition, during that time late 1800’s and early 1900’s bull rings (syndicates) were formed in Europe and the U.S. to allow dairymen the opportunity to afford and share top sires that had traits they wanted to add to their herds.
During World Wars I and II, European genetic improvement was disrupted. Some records were lost and destroyed and the rebuilding efforts disrupted progress while the U.S. started to move ahead with implementation of A.I. The advent and implementation of A.I. in the U.S. by the cooperative extension service provided the vehicle to distribute great sires. The first Artificial Insemination station using fresh semen was started in New Jersey in 1938. This allowed many farmers to breed more offspring through semen from top pedigreed bulls. Frozen semen was developed by Dr. Christopher Polled in England in 1951. This allowed for the storage and transportation of semen to greater distances and for greater amounts of time.
The major improvement was finally to be realized with the development of herdmate comparisons using Dr. Lush’s idea and implemented into progeny test programs. In 1962 herdmate comparisons were implemented. The first bull and cow indexes were published in 1964 by R.H. Miller. The concepts of linear type was developed by Dr. Paul Miller at Cornell in 1971 and adopted by the Jersey and Guernsey breed associations in 1980. This replaced the previous true-type model that was previously used by most of the associations.
Below you will see the trends in cow breeding values from 1957 through 2001. You can see that progress was slow until the advent of progeny testing that was implemented in the 1960’s. Since that time, improvements have been made in the area of evaluation models and increased number of sires to be selected and have allowed the continual increase in the genetic trend for milk production in the U.S.
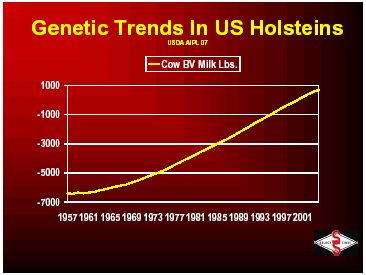
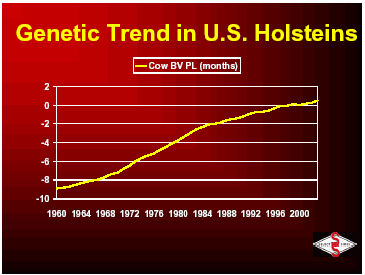
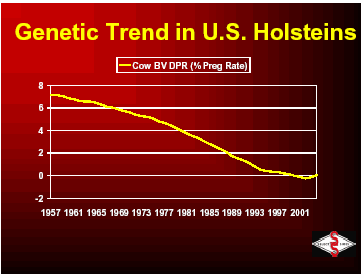
With the ability to breed for high production, more and more dairymen were requesting animals that could withstand the stress of high production, thus type traits were developed by breed associations to improve udders, feet, legs and total conformation. Low heritability of health traits made those unpopular as a breeding goal but was developed in the late 1990s by USDA. Today’s goal for Select Sires and many organizations around the industry is to develop a higher producing animal that is resistant to the many reasons that it would be culled from the herd. The United States Department of Agriculture and the breed associations are measuring over 29 different traits that are available for producers to use to improve the productivity and longevity of US dairy cattle.
However, today there are many challenges to the traditional progeny testing methods in providing genetic evaluations for elite males and females to make improvement and provide elite genetics. The first is the 5-year generation interval which takes a long time to create progress. Progeny testing is costly as to produce a good bull, only one out of 13 bulls progeny tested is selected for active service at Select Sires. In addition, there is slow progress due to low accuracy in low heritability traits that are usually associated with fitness. The new tools of molecular genetics show great promise to create some solutions to improve the speed and success of genetic development for AI companies and US herds to increase the rate of genetic progress by identifying those bulls who received a favorable sample of genes from their parents. So, let me take you through a review of the definitions of molecular genetics and discuss the possibilities.
The present practice of traditional progeny testing is to create a group of progeny (usually around 100 progeny of an animal), measure that progeny’s performance and compare it against its contemporary herdmates. This information is to estimate the parents’ contribution and create breeding values for the parents of those individuals. To select the animals to test, it is assumed that the genes of that individual are an equal distribution of half from the sire and half from the dam.
Molecular genetics is where we actually investigate, once the embryo is developed, the actual genes that were transmitted from the sires and dams of these individuals. Today, single nucleotide polymorphisms (SNP’s) have been mapped in the bovine by the U.S. Department of Agriculture through partnerships with many universities and countries around the world. These nucleotides make up the two strands of DNA that make up a chromosome. Chromosomes come in pairs and there are 30 pairs of chromosomes in each bovine. Where we find a single nucleotide on one strand of DNA that is different from normal, it is called a polymorphism. Polymorphism in its simplest term means different. These differences are then mapped and appear on various spots across the chromosome. The ability to track these differences and correlate their effects with known genetic levels will allow statisticians to develop prediction models to forecast the performance of an animal using these SNPs as they occur across the 30 chromosomes. Thus, the differences between full siblings can theoretically be identified as soon as the animal is created. Therefore, you will be able to predict the differences among various individuals in terms of their estimated transmitting abilities just from these molecular predictions. This information, when combined with the pedigree information, can be used to better predict the transmitting ability of that individual.
In 2001, a Dutch researcher named Meuwissen published a theoretical research study identifying the improvements that can be made using molecular markers to enhance the accuracy of prediction. As you will see in the chart below, the improved accuracy will move a high heritability trait from a pedigree of 40% to an estimated 85% with the additional marker information and a low heritability trait that is .25 to .30 will increase the accuracy of the estimated transmitted ability to .65 to .70.
| High Heritability | Low Heritability | |
|---|---|---|
| Pedigree | 0.40 | 0.25 - 0.30 |
| Ped + Markers | 0.80 - 0.85 | 0.65 - 0.70 |
|
Meuwissen, et al., 2001 (Genetics 157: 1819-1829)
|
||
In 2007, Dr. Paul VanRaden did a theoretical simulation using 50,000 SNPs equally distributed across the 30 chromosomes. He identified that the parent average reliability in the pedigree of 36% would increase to 58% to 71% in accuracy making major improvements in the accuracy of that estimate. Improved statistical methodologies such as a Baysian method could increase the reliability of these predictions as these statistical techniques are refined.
Under the direction of Dr. Curt VanTassel, the USDA’s Animal Improvement Laboratory is presently undergoing a study in cooperation with NAAB members who participate in the Cooperative Dairy DNA Repository (CDDR). In this research study, 3500 proven animals will be evaluated based on approximately 54,000 different SNPs in Holstein, Jersey and Brown Swiss. Each of these animals will be used to create a haplotype map. The scientists at USDA will develop statistical methodologies to compare their proven information with these SNPs and develop an estimation model for use in these breeds. This information could become part of the national genetic evaluations in early 2009. The contributors to the cooperative dairy DNA repository are Select Sires, CRI, ABS, Accelerated Genetics and Semex.
So, what should we expect to receive from this new marker information in regards to PTA’s? First, the PTA’s that would be presented would look just like those presented today in either the form of a progeny proof or a parent average PTA. The difference is that the reliability will be higher when the molecular evaluations have been preformed. Conservative initial estimates of Increases in accuracy of production and type estimations of male and female PTA’s will go from 30 to 40% in reliability based on the trait to an average of 45 to 60% reliability for most production and type traits. In addition, major improvements in lower reliability traits such as productive life and DPR will move accuracies of a parent average from 15-30% to 40-55% reliability based on the trait. Thus, with the new tools we will know more about a virgin heifer who has had a molecular test, than we will about her mother with two calves and two lactations with no molecular testing. This information will be available in both Jerseys and Holsteins. In addition, as researchers have more time to develop more sophisticated statistical models, we can expect the accuracies of each of these traits to improve over time.
The benefits of this technology will be a better selection of bull dams and young bulls for progeny testing. This will increase the rate of genetic progress especially in improving low heritability traits. Dairy herds will be able to use young bulls with more confidence and less disaster where animals have serious negative assortment of genes. Additionally, dairymen will be able to better select dams from which to flush or save bulls for breeding as well. Faster progress in health traits will be important in all of the major dairy breeds as herd sizes and production levels continue to increase.
One area that has been talked about in major publications is inbreeding. Understanding the sources of genes will give us a better understanding of the level of inbreeding. This tool will allow us to slow or reduce the levels of inbreeding in production herds. The end result is that we should have better tools to make faster progress, higher producing, healthier, long-lived cows, and continue to expand the US influence in genetics both in the US and around the world.
Sources for this presentation: 100 Years of Research and Inquiry by the American Dairy Science Association; Meuwissen, et al., 2001 Genetics 157; 1819-1829; Paul VanRaden, PhD, USDA 2007, Dr. Kent Weigel, PhD, University of Wisconsin – Madison; Mr. Chuck Sattler MS, Vice President Genetic Programs, Select Sires, Inc.
Further Reading
| - | You can view a full list of the 45th Dairy Conference Production proceedings by clicking here. |
April 2008


